SARS-Cov-2 - part 3 - nsp1: A hopefully more detailed analysis of the cellular saboteur
Co-authored by Gabriella Jonasson
My German Cartesian mind (here we go with stereotypes) asks me to start with the first protein expressed by the viral genome of SARS-Cov-2, nsp1. That is, by far, not the low hanging fruit. But it is interesting to learn more about the roles of the different constituents of the virus. Nsp1 actually also has a few very interesting roles to facilitate the viral life-cycle in its host environment.
Before going into all the details, let me briefly explain how I’m going to structure this post and the upcoming ones. I’ll try to gather & summarise information I can find on the role, structure and the potential use of nsp1 as a therapeutic target, or at least what should be done to know whether it could be a therapeutic target one day. In order to evaluate these aspects I’ll organise the post in three parts. Firstly, I look at the sequence and its variability. Secondly, I’ll write a bout the protein’s role and go into more detail than what we usually can find in summaries like the one I wrote before. Lastly, I’ll list structures that could be used today, structures that could be of interest for homology modelling or structures that would be really interesting to obtain. In this last part, I’ll also discuss experiments to run and therapeutic options.
Sequence
Let’s first compare the protein sequence of the nsp1 region in SARS-CoV-2 with SARS-CoV.
Here’s a MUSCLE alignment of the two nsp1 sequences, using the SARS-CoV-2 as reference:
Alignment of SARS-CoV-2 nsp1 and SARS-CoV nsp1. Alignment done with MUSCLE and image generated with JalView.
The first information that can be extracted from this alignment is that for both SARS viruses the nsp1:s are very close. No large inserts or deletions are observed. Mutations observed are mainly amino acid substitutions of similar types (a part from a few exceptions).
The role of nsp1 is believed to be very similar in SARS-Cov and SARS-Cov-2. The mutations observed between both sequences here are likely not relevant for the functions of nsp1. I’ll try to complete this alignment with a statistical analysis on all nsp1’s of all different SARS-Cov and SARS-Cov-2 strains. It’ll allow to gain a bit more perspective on which amino-acids are important in the protein itself.
Role of the Protein
To better understand the role of nsp1 in SARS-CoV-2, let’s look at what’s known for two closely related coronaviruses - MERS-CoV and SARS-CoV.
MERS-CoV
The role of the MERS nsp1 has been described as Specific recognition of viral RNA that is required for efficient viral replication; possibly interacts with cyclophilins and is thought to be a major virulence factor because it suppresses protein synthesis through the degradation of host mRNA.
SARS-CoV
In SARS-CoV, nsp1 is described to suppress the host gene expression as well (including type I interferons). The authors of this study identified K164 and H165 as being important for the functional role of nsp1. They generated a K164A and H165A mutant and observed that gene expression of host mRNA was increased compared to the host mRNA levels (to some extent) with the wild-type virus. I highlighted both residues in the sequence alignment with a purple indicator - no difference between SARS-CoV and SARS-CoV-2.
Another contribution used mutagenesis to analyse the functional features of nsp1 and their role in host gene expression inhibition in a more systematic way. Here the authors followed the protein expression of nsp1 and the host mRNA gene expression but also looked at the inhibition of cell signalling, which is also inhibited by nsp1.
The set of mutations performed by the authors are all rather violent, but judge by yourselves:
Results from a systematic mutational study on SARS-CoV nsp1 reported here.
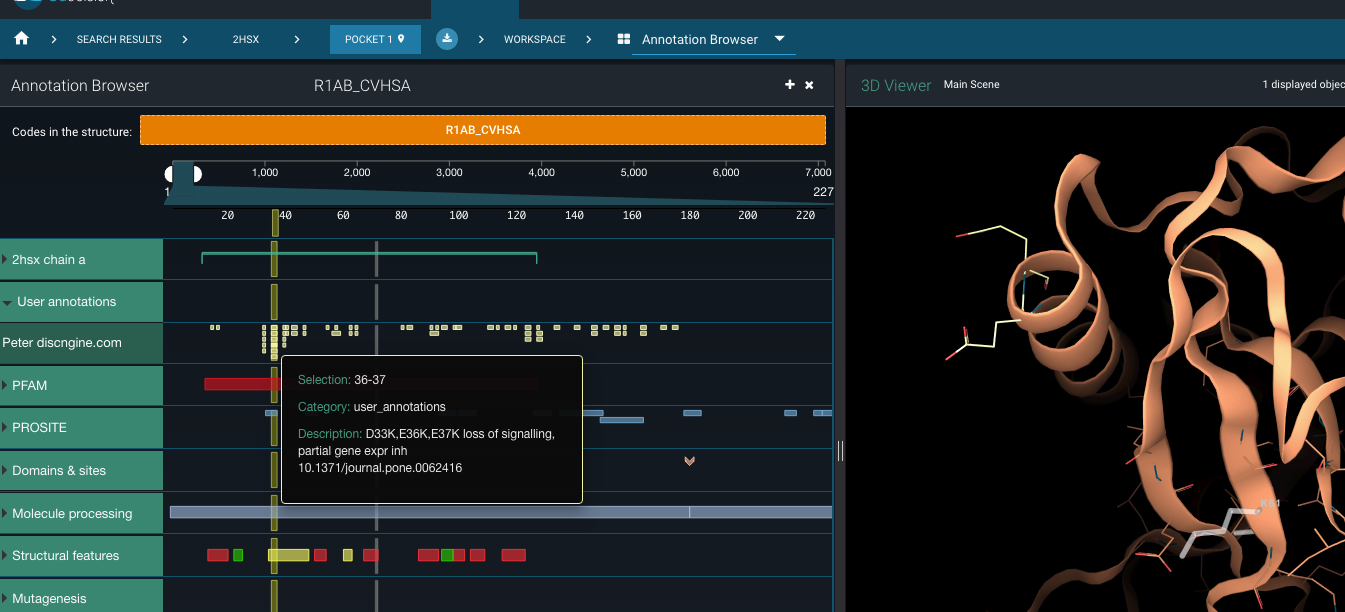
All of these mutations are available as sequence annotations on the SARS-CoV proteome in 3decision (UniProt code R1AB_CVHSA). For instance, if you visualise the SARS-CoV nsp1 structure 2hsx in 3decision, you’ll have access to the mutations’ positions on the 3D structure . Careful though, the quality of this particular structure is more than questionable.
After the last outbreak, efforts were made to resolve 3D structures of the SARS-CoV proteins. As a result, the 3D structure of nsp1 of SARS-CoV is actually suspected to have a beta barrel fold and two disordered (unstructured) regions, at the N and C terminal, respectively. An NMR structure of the “structured” part of nsp1 is available on RCSB and here on 3decision.discngine.cloud. However, this NMR structure is likely to be of low quality and exposes several Ramachandran outliers and unusual side-chain contacts. It’s therefore questionable to use this structure as a template to construct a homology model of SARS-CoV-2 nps1.
It is interesting to note that when using the alpha-fold based tr-rosetta in de-novo structure prediction, a fold different to the structure available of SARS-Cov nsp1 is found (for what it’s worth). Also, all other models I could spot so far were low quality and low confidence models for the SARS-Cov-2 nsp1.
I should also mention that I could not find any information about the quaternary state of the functional nsp1.
SARS-CoV-2
How can we use this information to better understand the role and structure of SARS-CoV-2 nps1?
The structured part of nsp1
In my opinion, it is rather difficult and risky to use the structural information on nsp1 available today. Unfortunately, the mutagenesis data available above does not really help improve this fact. So before being able to use nsp1 in a structure-based drug design effort, significant work is required to obtain reliable information.
The unstructured part of nsp1
But what about the role of unstructured parts of nsp1? The unstructured C-terminal domain is rather long and not modelled in the NMR structure SARS-CoV nsp1 (2hsx). This region ranges from amino acid 128 to 180, thus covering the positions 164-165, mentioned earlier, where mutations seemed to impact the gene expression of host mRNA.
When analysing the unstructured C-ter sequence in SARS-CoV-2 using hhpred (method described here, here, here and here), a single remote homolog (sub-hit) is found (see image below). Even though the relevance of this hit can be discussed (the matching sequence, amino acid 128-135, is very short), it happens to be chain D of the structure 3gme (visualise in 3decision or RCSB). This structure is a complex between the polynucleotide phosphorylase (PNPase) and the Ribonuclease E (RNase E) from E. coli. More precisely, the C-ter of SARS-CoV nsp1 matches the C-ter of the E-coli RNase E (visualise motif highlighted in 3decision). In this E-coli protein complex, the matching C-ter motif is structured as a beta strand and binds to PNPase by extending one of the PNPase’s beta sheets. This observation is interesting because the RNase E C-ter motif is predicted to be a coil - as the C-ter of nsp1 is described to be an intrinsically disordered region. We can, therefore, not rule out that the unstructured parts of nsp1 won’t bind in a structured way to other proteins.
Unique hit obtained via hhpred using “NKGAGGHSYGADLKSFDLGDELGTDPYEDFQENWNTKHSSGVTRELMRELNGG” (C-ter of nsp1 from SARS-CoV-2) as query.
My next question was: Did the authors of the E-coli protein complex structure (3gme) express and use the full RNase E to get their crystals? As a matter of fact, no they did not. They used a “microdomain” as they call it. So again it sheds some uncertainty to the relevance of this structure. However, the authors of this structure also highlight another very interesting fact in their paper - the complex they observe in E. coli is remarkably similar to an interaction in the human exosome complex. More precisely, it’s similar to the complex between the human RNase PH-like subunits Rrp45 and Rrp46. The C-terminal tail of Rrp46 plays the analogous role of the E-coli RNase E micro-domain and forms a pseudo-continuous β-sheet with the Rrp45 subunit.
So that’s interesting indeed. Let’s hypothetically consider that the SARS-CoV-2 nsp1 C-ter could interact at this precise location with Rrp45 and alter the interaction with the endogenous Rrp46 and thereby disrupt the function of the exosome complex. How would this effect the host cell?
In order to answer this question, let’s look closer on what the human exosome complex actually is, what it’s doing and where it comes from.
First of all, the exosome complex (or RNA exosome) is not to be confused with the the extracellular vesicles exosomes (confusing, I know). The exosome complex is an evolutionary conserved multi-protein complex important for the cleavage and degradation of RNA (further reading 1, 2, 3, 4, 5, 6. The ring-shaped core of this complex is composed of six RNase proteins (including Rrp45 and Rrp46), and three additional proteins with a RNA binding domain (see image below). This paper describes a few diseases that appear when something is awry with the exosome complex.
Structure of the yeast RNA Exosome complex (source). Spot the Rrp45/46 complex in the middle (yellow/red).
Overview of the interaction between Rrp45 (green) and Rrp46 (orange). Open the same view in 3decision here. Based on RCSB structure 2nn6
So back to our question. What would happen if nsp1 could bind to Rrp45? When looking at the 3D structure of the human exosome complex (see image above), it becomes clear that the sub-units Rrp45 and Rrp46 not only interact through the extension of the beta-sheet, but also build up a beta-sheet sandwich together. Their side-chains are of importance here. We can also observe that, the fold of Rrp45 is comparable to the fold of E. coli PNPase.
Next, the NMR structure of SARS-CoV nsp1 mainly contains a small beta-barrel-like fold. What if nsp1 is actually capable of mimicking the interaction of Rrp46 with Rrp45? It would alter host RNA processing and degradation. Interestingly, that’s one of the roles that are currently described for nsp1 from SARS-CoV & SARS-CoV-2.
Let’s check out if something could back-correlate with the mutation data we currently have at hand to check out if mutations have been tested on the small (possible) beta-strand interacting with Rrp45 (I’m building up hypotheses here, so nothing is verified scientifically, yet).
Within the amino acid 128-135 range of nsp1 (the hhpred match), we find the mutations m2, m4 and m35.
m35 alters H134D and S135Q. By analogy to the E coli structure and the hhpred hit we had before, residues 134 and 135 are not part of the interacting beta-strand but located in a loop on top of the beta-sheet. In the human Rrp45/Rrp46 complex this loop is forming a small helix as you can see in the forefront of screenshot above. m2 and m4 on the other hand affects resiudes 128, and 124, 125, 128 & 129, respectively. These are positioned within the range of what could be an Rrp45 interacting region of nsp1 (again, this is just an hypothesis)
m2, m4 and 35 lower the capability of nsp1 to inhibit RNA expression of the host according to the mutation study cited previously as well.
Let us quickly double check with the Krogan paper that came out recently (cited in my previous post) if we can somehow strengthen this hypothesis. They observed DNA polymerase alpha proteins interacting with nsp1. How does this explain the phenotype of reduced gene expression or immune response of the cell? … still a bit of reading to do here on my side.
Hypothesis to be validated experimentally:
As a result, it could be hypothesised that nsp1 interacts directly with Rrp45 at the location described in the session above (structure 2nn6).
Experiments that should be done to verify this hypothesis:
isolate Rrp45 and nsp1 and verify binding
if crystals of the human RNA exosome have been obtained in a lab before, could some be obtained with Rrp45 and nsp1?
3D models that should be built to design directed mutagenesis experiments:
complex of Rrp45 and nsp1 (not trivial as the nsp1 structure is very debatable and part of the C-ter is missing)
Additional roles of nsp1
Changing the structure of Nuclear Pore Complex
SARS-CoV nsp1 is known to associate with Nup93 and displaces it from the nuclear pore complex (NPC) assembly. The NPC is responsible for protein transport from the nucleus to the cytoplasm. Nup93 delocalisation is dependent on the presence of nsp1 in the cell. It is, however, unclear whether this Nup93 delocalisation is due to a direct interaction with nsp1 or via a mechanism not yet understood.
In this paper, it is also shown that an RNA binding protein, nucleolin, has been relocalised to the cytoplasm. These results offer a new mode of function by nsp1 in suppressing host cell’s function. I’ll come back to nucleolin by the end of this post, but that’s a common observation for several viral infections and a very intriguing one.
Stalling ribosome assembly & mRNA translation
Using a 2-pronged strategy to dampen host gene expression, nsp1 binds to the 40S ribosome at the 5′-untranslated region (UTR) of host mRNA and stalls further ribosome assembly, ultimately inhibiting host protein synthesis (source).
Furthermore, the nsp1-40S ribosome complex induced the modification of the 5'-region of capped mRNA template and renders the template RNA translationally incompetent. There are existing structures of the 40S ribosome and pre-ribosome complexes and it would be very interesting to obtain a high-quality structure of an nsp1, ribosome and mRNA complex.
This earlier paper describes are more complex mechanism of action of SARS-Cov nsp1 (very likely that the one of SARS-Cov-2 is very similar in function): “nsp1 inhibited the translation initiation step by targeting at least two separate stages: 48S initiation complex formation and the steps involved in the formation of the 80S initiation complex from the 48S complex. nsp1 had a differential, mRNA template-dependent, inhibitory effect on 48S and 80S initiation complex formation.”
Experimental structure or high-quality model needed:
Human 40S subunit with nsp1 bound to mRNA (structure uncertain).
Human 48S with nsp1
Several ribosome IRES complexes are resolved today, so this should be doable ;)?
Cleaving host mRNA
Furthermore, nsp1 seizes an unknown cellular endonuclease to cleave mRNA at the 5′-UTR, facilitating rapid decay of the cleaved mRNA by exonucleases (sources 1, 2, 3, 4 and 5). Could this also be linked to the exosome as described above?
Therapeutic options
Identifying therapeutic options at this stage is a challenging or even impossible task for nsp1. But hypothetically speaking several options could become of interest once the mode of action of nsp1 and how exactly it’s doing what it’s doing become clearer.
Nsp1 is known to bind mRNA. How difficult / relevant and specific would it be to design and test siRNA’s targeting specifically nsp1?
If nsp1 is indeed binding to proteins of the exosome, what approaches could be used to avoid that mechanism? Small molecule and antibody approaches are very likely impossible against nsp1.
Nucleolin as antiviral target?
So if targeting the viral protein is complicated, how about host proteins? Some of the current antivirals that attract attention (hydroxychloroquine for instance) alter things on the host, not directly in viral proteins. Previously I mentioned high concentrations of nucleolin in the cell as a result of the action of nsp1 from SARS-Cov.
This paper states: ”nsp1 triggers the cleavage and degradation of a majority of host mRNAs while its own genomic RNA is protected, increased amounts of nucleolin in the cytoplasm may alleviate the function of nsp1 during host shutoff by differential binding to different RNA sequences. Understanding the role of nucleolin binding to various mRNA sequences, including the viral RNA in the presence of nsp1, would help to clarify the role of nucleolin in host shutoff.”
Furthermore here it is hypothesised that the N protein of SARS-Cov (1 again) is interacting with nucleolin directly.
All of this information spiked my interest in nucleolin and there are actually quite a few papers on nucleolin as oncology target, but also as a target for antivirals. In some viral infections (HIV for example, but also several others) it has been described to be expressed also at the cell surface and facilitate the attachment of the virus on the cell surface (together with its main interactant). Also for SARS-CoV, a second interactant, other than Ace-2 has been identified (not nucleolin though yet).
In influenza infections, nucleolin plays also a central role in viral life-cycle. It would be of great interest to clarify the role of nucleolin during the SARS-Cov-2 infection as well.
One of the papers previously mentioned also lists several molecules tested as antivirals (mainly cell-surface nucleolin). Nucleolin is also an oncology target with several known inhibitors. These have profound effects on cell functioning and possible cytotoxic effects. However, it’d be of interest to study the effect of these inhibitors in light of a SARS-CoV-2 infection as well to better understand the role nucleolin plays during the infection.
Experiments suggested:
Test known / approved anti-nucleolin inhbitors during SARS-Cov-2 infection in-vitro.
Further reading on nucleolin:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5620558/
https://www.tandfonline.com/doi/pdf/10.4161/rna.19718
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4560460/
Quadruplex & nucleolin: https://europepmc.org/article/pmc/pmc5466061
SARS Quadruplex - nucleolin (nsp3 related): https://europepmc.org/article/PMC/2674928
Supplementary information
Worth a read
https://www.sciencedirect.com/science/article/pii/S0166354213002234?via%3Dihub
https://www.fasebj.org/doi/abs/10.1096/fasebj.2019.33.1_supplement.649.7
Links to reference sequence data
SARS-Cov-2 nsp1 sequence was taken from here
SARS-Cov nsp1 sequence was taken from here
MERS-Cov nsp1 sequence was taken from here
Previous articles
SARS-CoV-2 - part 2 - From the viral genome to protein structures
SARS-CoV-2 - part 1 - Thriving for a systematic target and hit ID effort